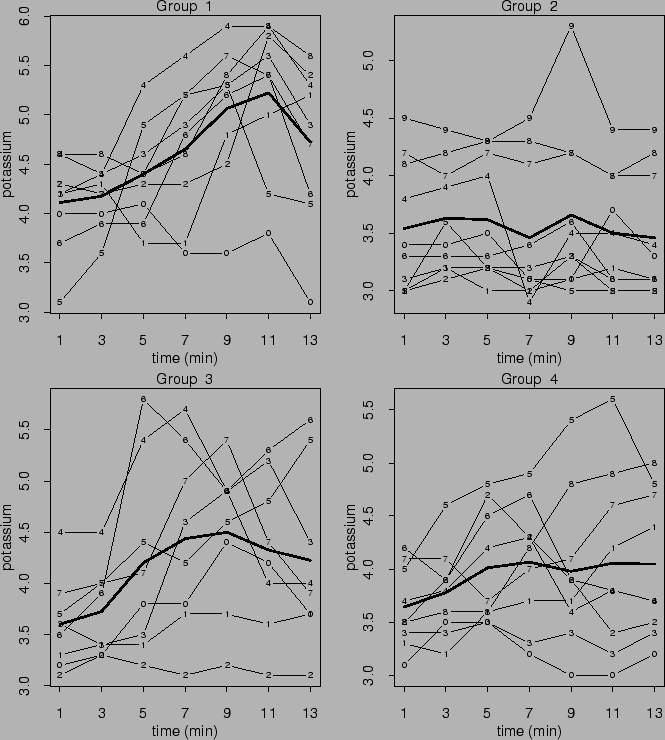
36 dogs were assigned to four groups: control, extrinsic cardiac
denervation three weeks prior to coronary occlusion, extrinsic
cardiac denervation immediately prior to coronary occlusion,
and bilateral thoratic sympathectomy and stellectomy three weeks
prior to coronary occlusion. Coronary sinus potassium
concentrations were measured on each dog every two minutes from
1 to 13 minutes after occlusion (Grizzle and Allen, 1969).
Observations are shown in Figure ![]() .
.
 |
We are interested in (i) estimating the group (treatment) effects;
(ii) estimating the population mean concentration as functions of
time; and (iii) predicting response over time for each dog.
There are two categorical covariates group and
dog and a continuous covariate time. We code
the group factor as 1 to 4, and the observed dog
factor as 1 to 36. We transform the time variable into
[0,1]. We treat group and time are fixed factors.
From the design, the dog factor is nested within the
group factor. Therefore we treat dog as a random
factor. For group ![]() , denote
, denote ![]() as the
population from which the dogs in group
as the
population from which the dogs in group ![]() were drawn
and
were drawn
and ![]() as the sampling distribution.
Assume the following model
as the sampling distribution.
Assume the following model
Suppose that we want to model the time factor using a cubic spline, and shrink both the group and dog factors toward constants. We define the following four projection operators:
![\begin{eqnarray*}
P_2f &=& \int_{{\cal B}_k} f(k,w,t) d{\cal P}_k(w), \\
P_1f &...
..._4f &=& [\int_0^1 (\partial f(k,w,t) / \partial t) dt] (t-0.5) .
\end{eqnarray*}](img695.png)
Then we have the following SS ANOVA decomposition
Based on the SS ANOVA decomposition (![]() ),
we will fit the following three models.
),
we will fit the following three models.
Model 1 includes the first seven terms in
(![]() ). It has a different population mean
curve for each group plus a random intercept for each
dog. We assume that
). It has a different population mean
curve for each group plus a random intercept for each
dog. We assume that
![]() ,
,
![]() ,
and they are mutually independent.
,
and they are mutually independent.
Model 2 includes the first eight terms in
(![]() ). It has a different population mean
curve for each group plus a random intercept and a random
slope for each dog. We assume that
). It has a different population mean
curve for each group plus a random intercept and a random
slope for each dog. We assume that
![]() ,
,
![]() ,
and they are mutually independent.
,
and they are mutually independent.
Model 3 includes all nine terms in
(![]() ). It has a different population mean
curve for each group plus a random intercept, a random
slope and a smooth random effect for each dog. We assume that
). It has a different population mean
curve for each group plus a random intercept, a random
slope and a smooth random effect for each dog. We assume that
![]() ,
,
![]() 's are stochastic processes which are
independent between dogs with mean zero and
covariance function
's are stochastic processes which are
independent between dogs with mean zero and
covariance function
![]() ,
,
![]() ,
and they are mutually independent.
,
and they are mutually independent.
Now we show how to fit these three models using slm.
Notice that the fixed effects, ![]() ,
, ![]() ,
,
![]() and
and ![]() are penalized.
Model 1 and Model 2 can be fitted easily as follows.
are penalized.
Model 1 and Model 2 can be fitted easily as follows.
> dog.fit1 <- slm(y~time, rk=list(cubic(time), shrink1(group),
rk.prod(kron(time-.5), shrink1(group)),
rk.prod(cubic(time), shrink1(group))),
random=list(dog=~1), data=dog.dat)
> dog.fit1
Semi-parametric linear mixed-effects model fit by REML
Model: y ~ time
Data: dog
Log-restricted-likelihood: -180.4784
Fixed: y ~ time
(Intercept) time
3.8716387 0.4339031
Random effects:
Formula: ~1 | dog
(Intercept) Residual
StdDev: 0.4980483 0.3924432
GML estimate(s) of smoothing parameter(s) : 0.0002334736 0.0034086209
0.0038452499 0.0002049233
Equivalent Degrees of Freedom (DF): 13.00377
Estimate of sigma: 0.3924432
> dog.fit2 <- update(dog.fit1, random=list(dog=~time))
> dog.fit2
Semi-parametric linear mixed-effects model fit by REML
Model: y ~ time
Data: dog.dat
Log-restricted-likelihood: -166.4478
Fixed: y ~ time
(Intercept) time
3.876712 0.4196789
Random effects:
Formula: ~ time | dog
Structure: General positive-definite
StdDev Corr
(Intercept) 0.4188372 (Inter
time 0.5593078 0.025
Residual 0.3403214
GML estimate(s) of smoothing parameter(s) :
1.674736e-04 3.286466e-03 5.778379e-03 8.944005e-05
Equivalent Degrees of Freedom (DF): 13.83273
Estimate of sigma: 0.3403214
To fit Model 3, we need to find a way to specify the smooth
(non-parametric) random effect ![]() . Let
. Let
![]() ,
,
![]() and
and
![]() .
Then
.
Then
![]() ,
where
,
where ![]() is the RK of a cubic spline evaluated at the design
points
is the RK of a cubic spline evaluated at the design
points
![]() . Let
. Let ![]() be the
Cholesky decomposition of
be the
Cholesky decomposition of ![]() ,
,
![]() ,
and
,
and
![]() .
Then
.
Then
![]() . We can write
. We can write
![]() , where
, where
![]() .
Then we can specify the random effects
.
Then we can specify the random effects
![]() using the matrix
using the matrix ![]() .
.
> D <- cubic(dog.dat$time[1:7])
> H <- chol.new(D)
> G <- kronecker(diag(36), H)
> dog.dat$all <- rep(1,36*7)
> dog.fit3 <- update(dog.fit2, random=list(all=pdIdent(~G-1), dog=~time))
> dog.fit3
Semi-parametric linear mixed-effects model fit by REML
Model: y ~ time
Data: dog.dat
Log-restricted-likelihood: -150.0885
Fixed: y ~ time
(Intercept) time
3.885269 0.4046573
Random effects:
Formula: ~ G - 1 | all
Structure: Multiple of an Identity
...
Formula: ~ time | dog %in% all
Structure: General positive-definite
StdDev Corr
(Intercept) 0.4671536 (Inter
time 0.5716811 -0.083
Residual 0.2383432
GML estimate(s) of smoothing parameter(s) :
8.775590e-05 1.560998e-03 3.346731e-03 2.870384e-05
Equivalent Degrees of Freedom (DF): 15.37699
Estimate of sigma: 0.2383432
We could use the anova function for linear mixed-effects
models to compare these three fits.
> anova(dog.fit1$lme.obj, dog.fit2$lme.obj, dog.fit3$lme.obj)
Model df AIC BIC logLik Test L.Ratio p-value
dog.fit1$lme.obj 1 8 376.9590 405.1307 -180.4795
dog.fit2$lme.obj 2 10 352.8955 388.1101 -166.4478 1 vs 2 28.06346 <.0001
dog.fit3$lme.obj 3 11 322.1771 360.9131 -150.0885 2 vs 3 32.71845 <.0001
So Model 3 is more favorable. We can calculate estimates of the population
mean curves for four groups as follows.
> dog.grid <- data.frame(time=rep(seq(0,1,len=50),4),
group=as.factor(rep(1:4,rep(50,4))))
> e.dog.fit3 <- intervals(dog.fit3, newdata=dog.grid, terms=rep(1,6))
Figure ![]() plots these
mean curves and their 95% confidence intervals.
plots these
mean curves and their 95% confidence intervals.
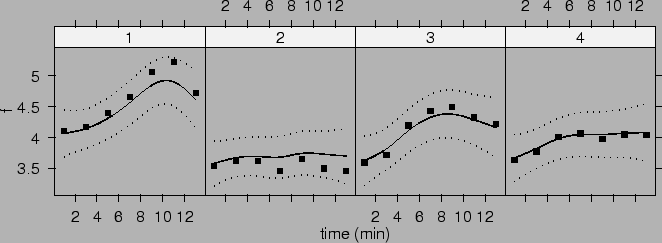 |
We have shrunk the effects of group factor toward
constants. That is, we have penalized the group main effect
![]() and the linear time-group interaction
and the linear time-group interaction
![]() in the SS ANOVA decomposition
(
in the SS ANOVA decomposition
(![]() ). From Figure
). From Figure ![]() we can see that
the estimated population mean curve for group 2 are
biased upward, while the estimated population mean curves for
group 1 is biased downward. This is because responses from
group 2 are smaller while responses from group 1 are larger
than those from groups 3 and 4. Thus their estimates are
pulled towards the overall mean. Shrinkage estimates in this case may
not be advantageous since group only has four levels.
One may want to leave
we can see that
the estimated population mean curve for group 2 are
biased upward, while the estimated population mean curves for
group 1 is biased downward. This is because responses from
group 2 are smaller while responses from group 1 are larger
than those from groups 3 and 4. Thus their estimates are
pulled towards the overall mean. Shrinkage estimates in this case may
not be advantageous since group only has four levels.
One may want to leave ![]() and
and
![]() terms
unpenalized to reduce biases. We can re-write the fixed
effects in (
terms
unpenalized to reduce biases. We can re-write the fixed
effects in (![]() ) as
) as
> dog.fit4 <- slm(y~group*time, rk=list(rk.prod(cubic(time), kron(group==1)),
rk.prod(cubic(time), kron(group==2)),
rk.prod(cubic(time), kron(group==3)),
rk.prod(cubic(time), kron(group==4))),
random=list(dog=~1), data=dog.dat)
> dog.fit4
Semi-parametric linear mixed-effects model fit by REML
Model: y ~ group * time
Data: dog
Log-restricted-likelihood: -173.0538
Fixed: y ~ group * time
(Intercept) group2 group3 group4 time group2:time
4.30592658 -0.70377899 -0.45507981 -0.54411601 0.70494987 -0.80352151
group3:time group4:time
-0.02604081 -0.33005253
Random effects:
Formula: ~1 | dog
(Intercept) Residual
StdDev: 0.4986658 0.3880457
GML estimate(s) of smoothing parameter(s) : 2.470589e-05 1.290422e+00
7.737418e-05 8.136673e-04
Equivalent Degrees of Freedom (DF): 13.05131
Estimate of sigma: 0.3880457
> dog.fit5 <- update(dog.fit4, random=list(dog=~time))
> dog.fit5
Semi-parametric linear mixed-effects model fit by REML
Model: y ~ group * time
Data: dog
Log-restricted-likelihood: -158.7129
Fixed: y ~ group * time
(Intercept) group2 group3 group4 time group2:time
4.31780496 -0.71566053 -0.46280282 -0.55513686 0.68418592 -0.78275742
group3:time group4:time
-0.01217737 -0.30620677
Random effects:
Formula: ~time | dog
Structure: General positive-definite, Log-Cholesky parametrization
StdDev Corr
(Intercept) 0.4225180 (Intr)
time 0.5536860 -0.005
Residual 0.3367037
GML estimate(s) of smoothing parameter(s) : 1.714698e-05 3.894053e+00
5.640272e-05 4.988060e-04
Equivalent Degrees of Freedom (DF): 13.74553
Estimate of sigma: 0.3367037
> dog.fit6 <- update(dog.fit5, random=list(all=pdIdent(~G-1), dog=~time))
> dog.fit6
Semi-parametric linear mixed-effects model fit by REML
Model: y ~ group * time
Data: dog
Log-restricted-likelihood: -142.3147
Fixed: y ~ group * time
(Intercept) group2 group3 group4 time group2:time
4.33679882 -0.72758015 -0.47372184 -0.57605073 0.65150749 -0.75063284
group3:time group4:time
0.00427347 -0.26053905
Random effects:
Formula: ~ G - 1 | all
...
Formula: ~time | dog %in% all
Structure: General positive-definite, Log-Cholesky parametrization
StdDev Corr
(Intercept) 0.4688705 (Intr)
time 0.5665381 -0.104
Residual 0.2394261
GML estimate(s) of smoothing parameter(s) : 8.175602e-06 1.567092e+00
3.081125e-05 1.289556e-01
Equivalent Degrees of Freedom (DF): 13.43655
Estimate of sigma: 0.2394261
> anova(dog.fit4$lme.obj, dog.fit5$lme.obj, dog.fit6$lme.obj)
Model df AIC BIC logLik Test L.Ratio p-value
dog.fit4$lme.obj 1 14 374.1076 423.0680 -173.0538
dog.fit5$lme.obj 2 16 349.4257 405.3804 -158.7129 1 vs 2 28.68192 <.0001
dog.fit6$lme.obj 3 17 318.6295 378.0814 -142.3148 2 vs 3 32.79624 <.0001
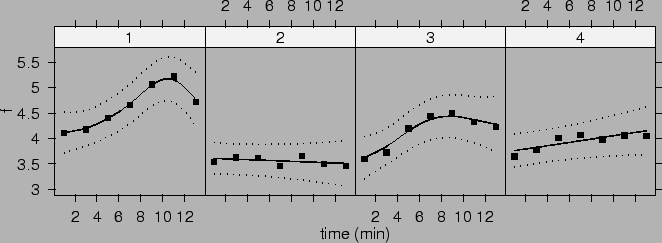
We note that the estimates are similar to, but not exactly the same as, those calculated by SAS proc mixed in Wang (1998b). The differences may be caused by different starting values and/or numerical procedures. We calculate estimates of the population mean curves for four groups as before.
> e.dog.fit6 <- intervals(dog.fit6, newdata=dog.grid, terms=rep(1,12))
Figure ![]() plots these mean curves and their 95% confidence intervals.
plots these mean curves and their 95% confidence intervals.
 |
We now show how to calculate predictions for dogs. Prediction
for dog ![]() in group
in group ![]() on a point
on a point ![]() can be computed as
can be computed as
![]() .
Prediction of the fixed effects can be computed using the
prediction.slm function.
.
Prediction of the fixed effects can be computed using the
prediction.slm function.
![]() and
and
![]() are provided in the estimates of the random
effects. Thus we only need to compute
are provided in the estimates of the random
effects. Thus we only need to compute
![]() .
Suppose that we want to predict
.
Suppose that we want to predict ![]() for dog
for dog ![]() in group
in group ![]() on
a vector of points
on
a vector of points
![]() . Let
. Let
![]() ,
,
> dog.grid2 <- data.frame(time=rep(seq(0,1,len=50),36),
dog=rep(1:36,rep(50,36)))
> R <- kronecker(diag(36),cubic(dog.grid2$time[1:50],dog.dat$time[1:7]))
> b <- as.vector(dog.fit6$lme.obj$coef$random$all)
> G.qr <- qr(G)
> c.coef <- qr.Q(G.qr)%*%solve(t(qr.R(G.qr)))%*%b
> tmp1 <- c(rep(e.dog.fit6$fit[dog.grid$group==1],9),
rep(e.dog.fit6$fit[dog.grid$group==2],10),
rep(e.dog.fit6$fit[dog.grid$group==3],8),
rep(e.dog.fit6$fit[dog.grid$group==4],9))
> tmp2 <- as.vector(rep(dog.fit6$lme.obj$coef$random$dog[,1],rep(50,36)))
> tmp3 <- as.vector(kronecker(dog.fit6$lme.obj$coef$random$dog[,2],
dog.grid2$time[1:50]))
> u.new <- as.vector(R%*%c.coef)
> p.dog.fit6 <- tmp1+tmp2+tmp3+u.new
Predictions for dogs 1, 2, 26 and 27 are shown in Figure ![]() .
.
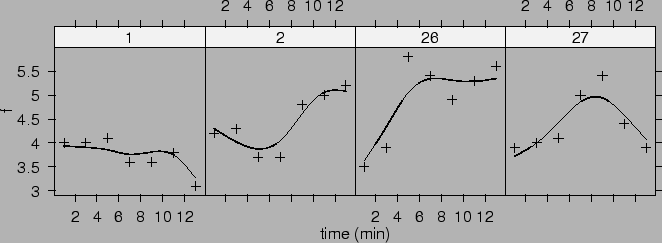 |
Observations close in time from the same dog may be correlated. In the following we fit a first-order autoregressive structure for random errors within each dog.
> dog.fit7 <- update(dog.fit6, cor=corAR1(form=~1|all/dog))
> dog.fit7
Model: y ~ group * time
Data: dog
Log-restricted-likelihood: -132.6711
Fixed: y ~ group * time
(Intercept) group2 group3 group4 time group2:time
4.343695660 -0.760617887 -0.483550117 -0.620851406 0.629970047 -0.715780104
group3:time group4:time
0.008927178 -0.239117441
Random effects:
Formula: ~ G - 1 | all
Structure: Multiple of an Identity
...
Formula: ~time | dog %in% all
Structure: General positive-definite, Log-Cholesky parametrization
StdDev Corr
(Intercept) 0.2779298 (Intr)
time 0.2909205 0.991
Residual 0.4456182
Correlation structure of class corAR1 representing
Phi
0.6359198
GML estimate(s) of smoothing parameter(s) : 2.921150e-05 1.954848e+06
1.098781e-04 1.893688e+01
Equivalent Degrees of Freedom (DF): 11.63503
Estimate of sigma: 0.4030084
By convention in nlme, model dog.fit7 may also be fitted as
dog.fit8 <- update(dog.fit6, cor=corAR1(form=~1))