Circadian rhythms have become a topic of intensive research during the last 40 years. Often the period is fixed as 24 hours due to the entrainment. However, for situations such as when individuals are denied exposure to zeitgebers or are living on irregular sleep/wake schedule, the period of the rhythm must be estimated from data. Several methods such as spectral analysis, autocorrelation, Enright's periodgram and linear regression of onset have been proposed in literature (Arendt et al., 1989; Refinetti, 1993). The first three methods require equidistant samples and the last method requires a marker for the onset of activity. All four methods requres several cycles of observations. Our method illustrated below, under the assumption that the shape of circadian rhythm remains unchanged, can be used for iregular design with few cycles.
This dataset is taken from a study on mood disorders. We thank
Daniel Buysse, MD, and Hernando Ombao, PhD, from the University
of Pittsburgh for permission to use part of the data. For several
healthy and depressed subjects, core body temperature (bt) is
measured over time every 5 minutes for 48 hours. Each subject
is put into an isolated room free of time cues. We only use
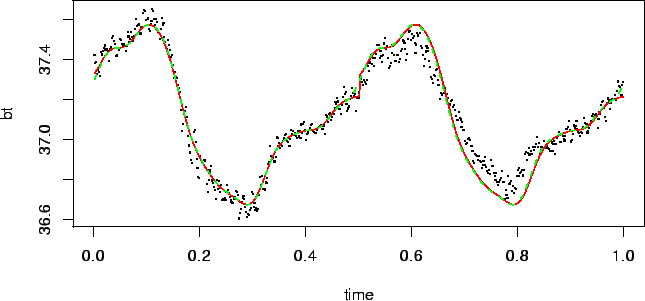
data from one subject and scale time variable into ![]() .
Measurements of this subject are shown in Figure
.
Measurements of this subject are shown in Figure ![]() .
The goal was to investigate possible effects of mode disorder
on the biological rhythms. For other physiological measurements,
it is known that biological rhythms exist. They are controlled
by the internal clock with a period close to 25 hours.
We will assume that biological rhythms exist for core body
temperature with a similar period. Then the 48 hour observations
contain at least one period. We need to estimate the period
and the shape function for each subject.
The common practice of fitting a sinusoidal function to each
subject may not be appropriate (Wang and Brown, 1996). We
demonstrate how to use SNR models to fit such data for
further analyses. See Hall et al. (2001) for another interesting
example and discussions on identifiability.
.
The goal was to investigate possible effects of mode disorder
on the biological rhythms. For other physiological measurements,
it is known that biological rhythms exist. They are controlled
by the internal clock with a period close to 25 hours.
We will assume that biological rhythms exist for core body
temperature with a similar period. Then the 48 hour observations
contain at least one period. We need to estimate the period
and the shape function for each subject.
The common practice of fitting a sinusoidal function to each
subject may not be appropriate (Wang and Brown, 1996). We
demonstrate how to use SNR models to fit such data for
further analyses. See Hall et al. (2001) for another interesting
example and discussions on identifiability.
 |
We consider the following model
> cbt.fit1 <- snr(bt~f(time-a**2*floor(time/a**2)),
func=f(u)~list(~1, periodic(u)),
params=list(a~1), data=cbt, spar="m",
start=list(params=c(sqrt(.5))), cor=corAR1())
> cbt.fit1
Semi-parametric Nonlinear Regression Model Fit
Model: bt ~ f(time - a^2 * floor(time/a^2))
Data: cbt
Log-likelihood: 1141.090
Coefficients:
a
0.7094294
Correlation Structure: AR(1)
Formula: ~1
Parameter estimate(s):
Phi
0.8456162
Smoothing spline:
GML estimate(s) of smoothing parameter(s): 6.161782e-08
Equivalent Degrees of Freedom (DF): 21.72078
Residual standard error: 0.06496144
Number of Observations: 576
Converged after 4 iterations
> p.cbt.fit1 <- predict(cbt.fit1)
We can also fit an equivalent model
> cbt.fit2 <- snr(bt~f(a**2*time), func=f(u)~list(~1, periodic(u)),
params=list(a~1), data=cbt, spar="m",
start=list(params=c(sqrt(2))), cor=corAR1())
> cbt.fit2
Semi-parametric Nonlinear Regression Model Fit
Model: bt ~ f(a^2 * time)
Data: cbt
Log-likelihood: 1143.045
Coefficients:
a
1.412535
Correlation Structure: AR(1)
Formula: ~1
Parameter estimate(s):
Phi
0.8471175
Smoothing spline:
GML estimate(s) of smoothing parameter(s): 4.377812e-07
Equivalent Degrees of Freedom (DF): 21.38732
Residual standard error: 0.0649923
Number of Observations: 576
Converged after 4 iterations
> p.cbt.fit2 <- predict(cbt.fit2)
Fits from these two models are shown in Figure